UMAP
Tables
In Situ Projection
Gene Differential Expression Tests
Tests run with across CellType, CellType (Predict), and Cluster and split by organism. The differential testing has been updated (yet again). This time
to a "pseudoBulk" approach where gene counts are summed by study and the test of interest (e.g. Cluster). The summed counts are "bulk-like"
in their statistical properties and we simply apply them to a DESeq2 based differential test where we use the study as a covariate.
Two types of contrasts are extracted: in Table 1 we show a comparison (e.g. Rod) against all other remaining (everything not rod) and in Table 2 we use a contrast run in a pair-wise manner
where we test, for example, cluster 2 directly against against cluster 10.
Haystack
singleCellHaystack
is a
cluster or cell type independent method for identifying differentially expressed or "interesting" genes
Very briefly, it uses the
DKL
divergence across the scVI multidimensional space to find non-randomly expressed genes. The table is ordered
by the log10(p value) calculated (lower is a lower p value). A higher D_KL score means that the genes is less randomly
expressed. T counts sums the number of counts (higher is expressed in more cells).
The CellType(s) and Cluster columns are the "top" genes which are most differentially expressed in the
comparison. The idea is to provide a quick way to see what CellType(s) or Cluster are driving the
singleCellHaystack identified gene.
Data
The codebase for the creation of the scEiaD dataset is on
github
If size not given, it is less than 1 GB
Run plae Locally
If you have 500GB (!) of free hard drive space, you can run plae on your own computer.
Installation instructions are available in our Github repository
(this is the codebase for the app you are using now).
Seurat Objects
AnnData Objects
PseudoBulk Diff Testing Results
PseudoBulk Data Matrices (and Metadata)
Metadata
Counts
plae v0.95
PLatform for Analysis of scEiad
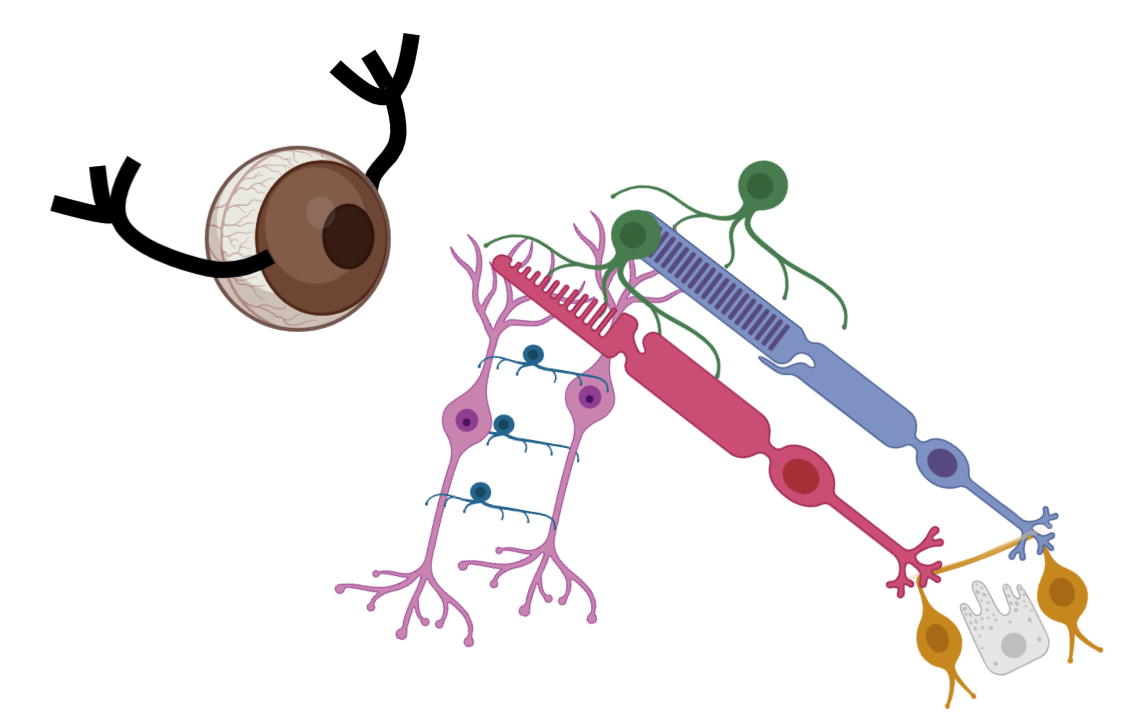
What is scEiaD?
single cell Eye in a Disk
The light-sensitive portion of the eye is the retina. The retina itself is not a monolithic tissue - there are over 10 major cell types. The cones and rods which convert light into signal are supported by a wide variety of neural cell types with distinct roles in interpretting and transmitting the visual signal to the brain. Behind the retina is the RPE and vasculature, which supports the high energetic needs of the rods and cones. In front of the retina is the clear lens and cornea, which shape the light onto the retina. scEiaD is a meta-atlas that compiles 1.1 million single-cell eye and body tissue transcriptomes across 44 datasets, 35 publications, and 4 species. Deep metadata mining, rigorous quality control analysis, differential gene expression testing, and deep learning based batch effect correction in a unified bioinformatic framework allow the universe of ocular single cell expression information to be analyzed in one location.
tldr
You can look up gene expression by retina cell type across loads of different studies, four organisms, and multiple developmental stages.
How to cite?
The article covering of the data creation and benchmarking of version 0.74 data is now publised at (now **not** on plae, but the codebase and principles are the same)
GigaScience!
Licensing
This work is released under the CC0 license
Data Sources
| Citation | PMID | SRA Accession | organism | Platform | Count | Labels |
|---|---|---|---|---|---|---|
| Yamagata M, Yan W, Sanes JR. A ... | 33393903 | SRP286543 | Gallus gallus | 10xv2 | 37498 | Yes |
| Collin J, Queen R, Zerti D, Bo ... | 33865984 | SRP275814 | Homo sapiens | 10xv2 | 100472 | Yes |
| He S, Wang LH, Liu Y, Li YQ et ... | 33287869 | SRP292721 | Homo sapiens | 10xv2 | 79245 | Yes |
| Lu Y, Shiau F, Yi W, Lu S et a ... | 32386599 | SRP223254 | Homo sapiens | 10xv2 | 64425 | Yes |
| Cowan CS, Renner M, De Gennaro ... | 32946783 | EGAD00001006350 | Homo sapiens | 10xv2 | 55018 | Yes |
| Lu Y, Shiau F, Yi W, Lu S et a ... | 32386599 | SRP151023 | Homo sapiens | 10xv2 | 42827 | Yes |
| Yan W, Peng YR, van Zyl T, Reg ... | 32555229 | SRP255195 | Homo sapiens | 10xv2 | 25737 | No |
| Voigt AP, Whitmore SS, Mulfaul ... | 32531351 | SRP257883 | Homo sapiens | 10xv3 | 24780 | Yes |
| Gautam P, Hamashima K, Chen Y, ... | 34584087 | SRP255012 | Homo sapiens | 10xv2 | 23610 | Yes |
| Sridhar A, Hoshino A, Finkbein ... | 32023475 | SRP238587 | Homo sapiens | 10xv2 | 18575 | No |
| Voigt AP, Mulfaul K, Mullin NK ... | 31712411 | SRP218652 | Homo sapiens | 10xv3 | 12634 | No |
| Ligocki A, Fury W, Gutierrez C ... | 34381080 | SRP362101 | Homo sapiens | 10xv2 | 12289 | No |
| van Zyl T, Yan W, McAdams A, P ... | 32341164 | SRP255871 | Homo sapiens | 10xv2 | 10858 | Yes |
| Patel G, Fury W, Yang H, et al ... | 32439707 | SRP254408 | Homo sapiens | 10xv2 | 10618 | No |
| Lukowski SW, Lo CY, Sharov AA ... | 31436334 | E-MTAB-7316 | Homo sapiens | 10xv2 | 9725 | Yes |
| Lu Y, Shiau F, Yi W, Lu S et a ... | 32386599 | SRP170761 | Homo sapiens | 10xv2 | 5002 | No |
| Menon M, Mohammadi S, Davila-V ... | 31653841 | SRP222958 | Homo sapiens | DropSeq | 3894 | Yes |
| Voigt AP, Whitmore SS, Flamme- ... | 31075224 | SRP194595 | Homo sapiens | 10xv3 | 3645 | Yes |
| Yan W, Peng YR, van Zyl T, Reg ... | 32555229 | SRP255195 | Homo sapiens | 10xv3 | 3187 | No |
| Swamy VS, Fufa TD, Hufnagel RB ... | 34651173 | SRP329495 | Homo sapiens | 10xv2 | 1544 | No |
| Menon M, Mohammadi S, Davila-V ... | 31653841 | SRP222001 | Homo sapiens | 10xv2 | 1273 | Yes |
| Voigt AP, Binkley E, Flamme-Wi ... | 32069977 | SRP238409 | Homo sapiens | 10xv3 | 1195 | No |
| Hu Y, Wang X, Hu B, Mao Y et a ... | 31269016 | SRP125998 | Homo sapiens | SMARTSeq_v2 | 8 | No |
| Peng YR, Shekhar K, Yan W, Her ... | 30712875 | SRP158528 | Macaca fascicularis | 10xv2 | 85327 | Yes |
| van Zyl T, Yan W, McAdams A, P ... | 32341164 | SRP255874 | Macaca fascicularis | 10xv2 | 4499 | Yes |
| Clark BS, Stein-O'Brien GL, Sh ... | 31128945 | SRP158081 | Mus musculus | 10xv2 | 127434 | Yes |
| Dani N, Herbst RH, McCabe C, G ... | 33932339 | SRP310237 | Mus musculus | 10xv2 | 64671 | Yes |
| Tabula Muris Consortium., Over ... | 30283141 | SRP131661 | Mus musculus | 10xv2 | 60587 | Yes |
| Tran NM, Shekhar K, Whitney IE ... | 31784286 | SRP212151 | Mus musculus | 10xv2 | 46175 | Yes |
| Yan W, Laboulaye MA, Tran NM, ... | 32457074 | SRP259930 | Mus musculus | 10xv2 | 44560 | No |
| Wu F, Bard JE, Kann J, Yergeau ... | 33674582 | SRP257758 | Mus musculus | 10xv2 | 42692 | No |
| Shekhar K, Lapan SW, Whitney I ... | 27565351 | SRP075719 | Mus musculus | DropSeq | 24158 | Yes |
| Heng JS, Hackett SF, Stein-O'B ... | 31843893 | SRP200499 | Mus musculus | 10xv2 | 15461 | No |
| van Zyl T, Yan W, McAdams A, P ... | 32341164 | SRP251245 | Mus musculus | 10xv3 | 14827 | Yes |
| Macosko EZ, Basu A, Satija R, ... | 26000488 | SRP050054 | Mus musculus | DropSeq | 12092 | Yes |
| Lehmann GL, Hanke-Gogokhia C, ... | 32196081 | SRP216903 | Mus musculus | 10xv2 | 9607 | No |
| Fadl BR, Brodie SA, Malasky M, ... | 33088174 | SRP269635 | Mus musculus | 10xv2 | 8516 | No |
| Balasubramanian R, Min X, Quin ... | 34757798 | SRP228556 | Mus musculus | 10xv3 | 8463 | Yes |
| Buenaventura DF, Corseri A, Em ... | 31260032 | SRP200599 | Mus musculus | 10xv2 | 8207 | No |
| Lo Giudice Q, Leleu M, La Mann ... | 31399471 | SRP168426 | Mus musculus | 10xv2 | 5040 | No |
| O'Koren EG, Yu C, Klingeborn M ... | 30850344 | SRP186407 | Mus musculus | 10xv2 | 3621 | No |
| Lo Giudice Q, Leleu M, La Mann ... | 31399471 | SRP186396 | Mus musculus | SMARTSeq_v2 | 599 | No |
| Clark BS, Stein-O'Brien GL, Sh ... | 31128945 | SRP158081 | Mus musculus | SMARTSeq_v2 | 505 | No |
| Fadl BR, Brodie SA, Malasky M, ... | 33088174 | SRP269634 | Mus musculus | 10xv2 | 358 | No |
| Shekhar K, Lapan SW, Whitney I ... | 27565351 | SRP075720 | Mus musculus | SMARTSeq_v2 | 337 | No |
| Shekhar K, Lapan SW, Whitney I ... | 27565351 | SRP073242 | Mus musculus | SMARTSeq_v2 | 246 | No |
scEiaD Curated Published Cell Type Labels
| CellType | Species | Studies | Count |
|---|---|---|---|
| Retinal Ganglion Cell | GG, HS, MF, MM | 11 | 68030 |
| Rod | GG, HS, MF, MM | 11 | 50622 |
| Fibroblast | HS, MM | 7 | 45697 |
| Amacrine Cell | GG, HS, MF, MM | 8 | 42005 |
| Epithelial | HS, MM | 4 | 35426 |
| Muller Glia | GG, HS, MF, MM | 13 | 34821 |
| Bipolar Cell | GG, HS, MF, MM | 12 | 33777 |
| Keratocyte | HS | 1 | 31850 |
| T/NK-Cell | HS, MF, MM | 7 | 29486 |
| Early RPC | MM | 1 | 25441 |
| RPC | HS, MM | 3 | 20899 |
| Late RPC | MM | 1 | 17645 |
| B-Cell | HS, MM | 4 | 16194 |
| Endothelial | HS, MF, MM | 12 | 12028 |
| Neurogenic Cell | HS, MM | 4 | 9219 |
| Rod Bipolar Cell | HS, MM | 2 | 9063 |
| Horizontal Cell | GG, HS, MF, MM | 7 | 8152 |
| Cone | GG, HS, MF, MM | 8 | 8040 |
| Mesenchymal | MM | 1 | 7840 |
| Macrophage | HS, MF, MM | 6 | 7190 |
| Photoreceptor Precursor | HS, MM | 4 | 6643 |
| Keratinocyte | HS, MM | 2 | 6272 |
| Melanocyte | HS, MF, MM | 8 | 6143 |
| Pericyte | HS, MF, MM | 7 | 5470 |
| Basal Cell | HS, MM | 2 | 5153 |
| Beam | HS, MF, MM | 3 | 4955 |
| Proliferating Cornea | HS | 1 | 4184 |
| Blood Vessel | HS | 2 | 4126 |
| Neural Crest | HS | 1 | 4030 |
| Smooth Muscle Cell | HS | 2 | 4005 |
| Schwann | HS, MF | 4 | 3225 |
| Monocyte | HS, MM | 4 | 2654 |
| Uveal | MM | 1 | 2578 |
| Red Blood Cell | HS, MM | 4 | 2507 |
| Satellite Cell | HS | 1 | 2295 |
| Corneal Progenitor | HS | 1 | 2243 |
| Corneal Epithelial | HS, MM | 3 | 2133 |
| Limbal | HS | 1 | 2040 |
| AC/HC Precursor | HS, MM | 3 | 1812 |
| Ciliary Margin | HS, MM | 2 | 1777 |
| Enterocyte | HS | 1 | 1725 |
| Plasma Cell | HS | 1 | 1591 |
| Ciliary Body | HS | 1 | 1476 |
| Ciliary Muscle | HS, MF | 2 | 1373 |
| Bladder | MM | 1 | 1191 |
| Bladder Urothelial | MM | 1 | 1154 |
| Mesenchymal (Stem) | MM | 1 | 1129 |
| Hepatocyte | MM | 1 | 1016 |
| Conjunctival Epithelial | HS | 2 | 914 |
| Corneal Endothelial | HS | 1 | 906 |
| Choriocapillaris | HS | 1 | 892 |
| JCT | HS, MF, MM | 3 | 821 |
| Mesoderm | HS | 1 | 718 |
| Microglia | HS, MF | 6 | 641 |
| Secretory Cell | HS | 1 | 352 |
| Corneal Basement Membrane | HS | 1 | 316 |
| Astrocyte | HS | 1 | 277 |
| Cholangiocyte | HS | 1 | 244 |
| Corneal Nerve | HS | 1 | 227 |
| Schlemm's Canal | MF | 1 | 119 |
| Limbal Progenitor | HS | 1 | 116 |
| Oligodendrocyte | GG | 1 | 111 |
| Meningeal | MM | 1 | 60 |
| Kidney Proximal Tubule | MM | 1 | 57 |
Labelled cell types from published papers were pulled, where possible, from a combination of the Sequence Read Archive (SRA), lab web sites, and personal correspondence, then adjusted to be consistent (e.g. MG to Muller Glia) between all studies. Only cell type - study combinations with >50 cells were included in this table.
scEiaD Machine Learned Cell Type Labels
| CellType | Species | Studies | Count |
|---|---|---|---|
| Amacrine Cell | GG, HS, MF, MM | 21 | 134443 |
| Rod | GG, HS, MF, MM | 22 | 119666 |
| Retinal Ganglion Cell | GG, HS, MF, MM | 21 | 107407 |
| Fibroblast | HS, MM | 16 | 74758 |
| Bipolar Cell | GG, HS, MF, MM | 25 | 67880 |
| Muller Glia | GG, HS, MF, MM | 20 | 62042 |
| Epithelial | HS, MM | 7 | 55571 |
| RPC | HS, MM | 9 | 43465 |
| Keratocyte | HS | 2 | 36650 |
| Late RPC | HS, MM | 7 | 34663 |
| T/NK-Cell | HS, MF, MM | 10 | 33876 |
| Early RPC | HS, MM | 10 | 32412 |
| Neural Crest | HS | 1 | 25239 |
| Corneal Epithelial | HS, MM | 5 | 22814 |
| Cone | GG, HS, MF, MM | 20 | 21394 |
| Endothelial | HS, MF, MM | 22 | 20744 |
| B-Cell | HS, MM | 8 | 19994 |
| Neurogenic Cell | HS, MM | 8 | 17785 |
| Photoreceptor Precursor | HS, MM | 5 | 14770 |
| Horizontal Cell | GG, HS, MF, MM | 15 | 14092 |
| Pericyte | HS, MF, MM | 16 | 11312 |
| Rod Bipolar Cell | HS, MM | 5 | 10331 |
| Blood Vessel | HS, MM | 5 | 10245 |
| Macrophage | HS, MF, MM | 12 | 10042 |
| Schwann | HS, MF, MM | 7 | 9366 |
| Mesenchymal | MM | 1 | 8054 |
| Melanocyte | HS, MF, MM | 12 | 8030 |
| Keratinocyte | HS, MM | 2 | 6635 |
| Smooth Muscle Cell | HS, MM | 4 | 6172 |
| Beam | HS, MF, MM | 5 | 5741 |
| Microglia | HS, MF, MM | 17 | 5674 |
| RPE | HS, MM | 6 | 5583 |
| Basal Cell | HS, MM | 2 | 5474 |
| Proliferating Cornea | HS | 1 | 4905 |
| Conjunctival Epithelial | HS, MM | 4 | 4467 |
| AC/HC Precursor | HS, MM | 6 | 4223 |
| Monocyte | HS, MM | 6 | 3233 |
| Satellite Cell | HS | 1 | 3126 |
| Red Blood Cell | HS, MM | 6 | 2991 |
| Uveal | HS, MM | 2 | 2860 |
| Corneal Progenitor | HS, MM | 5 | 2678 |
| Plasma Cell | HS | 1 | 1962 |
| Enterocyte | HS | 1 | 1877 |
| Ciliary Muscle | HS, MF, MM | 4 | 1756 |
| Limbal | HS | 2 | 1690 |
| Ciliary Body | HS | 1 | 1595 |
| Mesenchymal (Stem) | MM | 1 | 1395 |
| Bladder | MM | 1 | 1308 |
| Hepatocyte | MM | 1 | 1294 |
| Astrocyte | HS | 2 | 1183 |
| Bladder Urothelial | MM | 1 | 1165 |
| Ciliary Margin | HS, MM | 3 | 963 |
| JCT | HS, MF | 3 | 723 |
| Corneal Endothelial | HS | 2 | 664 |
| Corneal Nerve | HS | 2 | 374 |
| Secretory Cell | HS | 1 | 340 |
| Cholangiocyte | HS | 1 | 327 |
| Kidney Proximal Tubule | MM | 1 | 72 |
| Limbal Progenitor | HS | 1 | 60 |
The labels above were used to create a machine learning modeled which was used to relabel all* cells in the scEiaD (*above a confidence threshold of 0.5). Only cell type - study combinations with >50 cells were included in this table.
Contact
If you have questions about scEiaD dataset or the plae application, please contact
David McGaughey, Ph.D
.
Otherwise the National Eye Institute's Office of Science Communications, Public Liaison and Education responds directly to requests for information on eye diseases and vision research in English and Spanish. We cannot provide personalized medical advice to individuals about their condition or treatment.
Phone: 301-496-5248 — English and Spanish
Mail: National Eye Institute
Information Office
31 Center Drive MSC 2510
Bethesda, MD 20892-2510
Change log
0.95 (2024-12-09): Fixed diff table database slow performance, removed deprecated setSliderColor
0.94 (2023-06-12): Added error handling for exp plot situation where user filters all the data out. Fixed bug in logic for exp plot category filtering
0.93 (2023-02-16): Added the ability to download the plot data for the UMAPs, Expression Plot, Dot Plot, and Heatmap. New quick analysis document added
here
which gives a quick workthrough about how to recreate the plot locally in R. Enhanced the "Pop Up" buttons with more information and GIFs showing some functionality of the app.
0.92 (2022-08-30): Default in Exp Plot now facets on Gene. Exp Plot facet_wrap now on "free_y" instead of "free" to save space.
0.91 (2022-07-30): Updated diff testing model to a pseudoBulk / DESeq2 based approach. Now run on a per-species basis. Added heatmap visualization which uses the DESeq2 pseudobulk differential experession changes. Fixed bug that removed RPC from the Expression Plot view as well as display zero expression studies. Expression Plot view has a new option which flips flops the facet / x axis from Gene to whatever the cell type label is (CellType, CellType_predict, cluster).
0.90 (2022-04-05): Updated scEiaD scVI model once more. We simplified pan species gene name alignment by removing "one to many" or
"many to one" name alignments. All expressed genes are still retained in the individual species (if not present in the gene name name merged matrix), but we are less aggressive about merging gene names across species as we discovered some edge cases where many genes were getting
merged into one and vice versa. This necessitated a new core gene <-> cell count matrix and thus a new scVI model. We also took the opportunity to add a new mouse retina development dataset from
Balasubramanian et al. and a new ocular compartment dataset from Gautam et al. We leverage the new(ish) scANVI scVI approach in which we use the community
cell type labels to subtly improve the scVI modelling of the cell types. We hope this is the final update (hah) before we submit this work.
0.85 (2022-02-15): Updated example analysis to match current data, fixed 508/a11y compliance issues with the document.
Added *study level* seurat and anndata objects to "Data".
Updated the large downloadable Seurat / anndata objects with intronic counts data (velocity?!).
Added "Other Resources" section for alternative resources for ocular transcriptomics.
0.84 (2021-11-17): Added large human cornea dataset from Collins et al. As this was a brand new human dataset with several new cell types, a new scVI model was built. Hence the new apperance of the UMAP view. Fixed a filtering bug in Exp Plot.
0.83 (2021-11-01): Fixed bug in metadata filter table loading that was messing up some of the plots in certain situations. Tweak click table column choice to include organism.
0.82 (2021-10-26): Cool feature! Now you can click the UMAP viz to get cell info!
0.81 (2021-10-25): Fix small bug in bindCache logic, improve exp plot plotting by retaining zero expression studies
0.80 (2021-10-22): MASSIVE update. Chicken data added. Brain choroid added. Trabecular meshword added. Cornea added. Human body tissues added. We now have over one million cells in this resource. Counts cleaned up with
DecontX
to remove (mostly) Rod gene contamination (e.g. Rho *was* everywhere).
singleCellHaystack
table added to Diff Testing. Updated cell filtering with a higher minimum gene count cutoff to improve overall quality. Fixed bug in gene selection where human
genes that mapped to multiple mouse genes were accidently removed. Improved scran-based differential gene expression testing with better parameters and added logFC calculations
to improve interpretability. Fixed bug in dotplot plot where filtered data had the incorrect denominator values.
0.74 (2021-08-12): Remove broken link, fix bug in Expression Plot that was making average expression far too low.
0.73 (2021-06-10): Allow for UMAP plot to show all values (filter now starts at >=1)
0.72 (2021-04-29): Added more content and a table organzation to the Info -> Analysis... section
0.71 (2021-04-14): scEiaD preprint on bioRxiv! Added a filter in the "exp plot" section to remove data points with user-selected (default 50) minimum cells. I was finding that data points (e.g. cell type - study) with low N would often have "outlier" results. Added a new section to the web page - Analyses (under the "Info" tab)!
0.70 (2021-03-22): New scEiaD built with corrected fastq file sets (potential bug in 10x bamtofastq tool resulted ina few datasets getting scrambled barcodes). Removed a macaque dataset (SRR7733526) with odd behavior (clustering in 2D UMAP space largely alone). Tweaked the UMAP 2D gene view with a darker "background" cell color scheme to reduce "over-emphasis" on cells with low expression of a gene. CPM replaced with counts as some odd behaviour was detected in some genes in the UMAP view where there was high "background" expression. Counts have more consistent behavior. Removed hard filter that tossed cells with >2500 detected genes.
0.60 (2021-02-08): New scEiaD built with more studies. Removed several retinal organoid datasets that had snuck in. Added a filter option for the diff searching to search, for example, one cluster directly against another cluster.
0.52 (2021-01-13): Adding "missing" genes (we had only retained genes which were expressed in all three species, which naturally led to many genes (some important, like OPN1MW) to be dropped. That has been fixed. Tweak "Expression Plot" dot size to prevent crazy tiny point sizes.
0.51 (2021-01-06): Hello 2021!
Adam Gayoso
kindly pointed out that I was using scVI in a non-optimal manner, so I updated the scVI modeling to match their recommend "scArches" parameters. This (fortunately for my sanity) only subtly changes the downstream result. The more significant change is that we have totally changed the diff testing section to use the scran findMarkers test instead of our complicated and compute expensive pseudo-Bulk testing which continually gave odd results.
0.50 (2020-12-31): Goodbye 2020! Major update to the scVI-based UMAP projection which improves data quality. Removed non-tissue samples (e.g. organoid/cell lines). They will be added back later once I figure out a logical/simple way to do it. Fixed major bug in QC filtering which failed to remove high mitochondrial count (likely apoptosing cells) cells. Dot plot tweaked to improve relative dot sizes. Cowan et al. dataset added.
0.43 (2020-11-09): Downloadable diff results added to "Data." The diff results reactive data table now has a "Download all ..." button which replaces the "CSV" button that only downloaded the viewable data (100 max).
0.42 (2020-10-16): Alt text added to each button, tweaked UMAP-Tables layout again. Slide logo added. Site went public at the version on 2020-11-02!
0.41 (2020-10-06): Download buttons added for each plot.
0.40 (2020-10-05): UI and text labels tweaked in UMAP-Tables to improve tab selection order. Dot plot given a bar plot to show category size. Error handling improved when user fails to provide a category value to filter on. Data Table help button added. Help buttons moved to bottom of page with consistent visual - tabbing order. Colors of UI elements tweaked to improve contrast.
0.39 (2020-09-18): Fixed calculation error in dotplot where expression not scaled by number of cells in grouping variable.
0.38 (2020-09-02): Contact section and footer added for compliance.
0.37 (2020-08-24): Help pop section populated with text. Put white halo back around text in UMAP - Meta section. Loading circles added to plots. Row names added to tables. Diff testing filtered to only return results with FDR < 0.05 and abs(logFC) > 0.5.
0.36 (2020-08-17): Data download section added. Change log moved to separate section. CSS tweaked to show links in blue. First overview table updated to improve contrast. UMAP plots axis fixed.
0.35 (2020-08-14): Moved Overview tables to html for improved rendering and switched over color-blind friendly palette. Temporarily removed Temporal Plotting section. Improved filtering for Facet Plot. DotPlot plotting fixed and improved. Back-end server.R code moved into separate functions. Colors fixed so they stay consistent when filtering/subsetting the plots. Site now starts from scratch in under 5 seconds with improved fst-based data loading and pre-calculating more operations.
0.34 (2020-08-03): Fixed issue with TabulaMuris labels not appearing. Scanned app with koa11y for 508 compliance - changed headers from h2 to h1 to comply.
0.33 (2020-07-30): Exp plot can now take space or comma separated Genes as input. User can selected number of columns in Exp Plot. Diff Table formatting improved with rounding and PB_Test can be selected as a drop down now in the data table search.
0.32 (2020-07-29): In situ Projection viz added courtesy of Zachary Batz! It's a simulated cross section of the retina with each cell type colored by intensity of scRNA expression! Move table draw button under filtering in UMAP - Tables. Sort Diff Exp results by FDR. Filtering on numeric column now returns slider UI. Remove super dangerous ability to create faceted plots on numeric values.
0.31 (2020-07-24): Re-created scEiaD with better internal (Hufnagel) transwell RPE labelling (there are roughly two groups - mature RPE with high TTR expression and less (?) mature RPE with lower TTR), removal of the SRP166660 study as it was *all* non-normal (injured retina) (confirmed with correspondence with Dr. Poche), removed the pan RGC CellType labelling for the SRP212151 as I see post-hoc that there are LOADS of non-RGC cells. Did the same for SRP186407, which has substantial non-microglia. Generally, FACS != 100% celltype purity. Added differential testing against all Tabula Muris cell types. Removing clusters/cells with high doublet scores. Added cell cycle phase (G1/G2M/S) assignment. More study level metadata.
0.30 (2020-07-20): Huge update. Hundreds of thousands of cells added. The Tabula Muris project data (pan mouse) has been added to faciliate non-eye comparison. Filtering options added to most of the plotting views to allow for quick slicing into this huge dataset. Differential expression testing totally reworked - now uses "pseudoBulk" approach to better utilize the large number of studies we have.
0.23 (2020-06-16): Remove low N cell type from diff expression tables, tweak Overview with spacing alterations and updated text.
0.22 (2020-06-15): Added expression plot by user selected groups plot view. Fixed bug in mean cpm expression calculation for Viz -> UMAP - Table gene tables
0.21 (2020-06-15): Added subcluster diff testing tables, temporal gene expression by celltype plot section.
0.20 (2020-06-06): New 2D UMAP projection that includes the full Yu - Clark Human scRNA dataset. Added tables to "Overview" section showing data stats. Added "filtering" functionality to UMAP plot section.
Other potentially useful resources for single cell ocular trancriptomics
Independent datasets
: uses data from multiple independent groups
Harmonization datasets
: integrates data from multiple resources with consistent bioinformatic tooling